Massina Abderrahmane, Brice Hoffmann, Nicolas Devaux, Maud Jusot
A Matter of Representation…
Peptides are a promising class of therapeutics with unique properties. They share some advantages of proteins, such as their high selectivity but also some of small molecules, like metabolic stability and their low immunogenicity [1,2]. They can occupy larger surfaces of interaction than drug-like compounds, reaching higher specificities and making them interesting for targeting protein-protein interactions [2]. Despite these remarkable properties, their synthesis, size and flexibility make peptides aside in the world of therapeutics.
Peptides are biomolecules composed of amino acid chain(s) presenting a wide range of length (up to around 50 residues), with a molecular weight fluctuating between 200 and 5000 Da. They present a high flexibility and an important number of hydrogen-bond donors (HBD) (because of peptide bonds) giving them properties outside of Lipinski’s rule of 5 (Ro5), a traditional metric used to predict the drug-likeness of a molecule [3]. For example, Ro5 suggests that molecules should have a molecular weight lower than 500 Da and a number of HBD that should not exceed 5. On the contrary, most peptides currently available as drugs weigh up to 1200 Da and have 5 times more HBD [3].
For many years, peptides have suffered from several drawbacks in the drug discovery field, with major barriers such as lack of oral bioavailability and poor membrane permeability. Currently, 90% of all peptide drugs are still delivered by injection. However, advances in chemistry to produce more stable peptides have been made in the last few decades, triggering a new interest for peptides as therapeutic drugs. A proposed solution to improve peptide properties is the use of chemical modifications such as N-methylations, D-amino acids or cyclization. These modifications can improve peptides’ stability, reduce their degradation by proteases, increase the membrane permeability and even modulate favorably the binding affinity [4]. In particular, cyclic peptides present remarkable properties: cyclization lowers the flexibility compared to linear peptides and improves metabolic stability. Different types of cyclic peptides exist. Some examples are presented in Figure 1.
Currently, more than 80 peptide drugs have reached the market for a wide range of diseases, including diabetes, cancer, osteoporosis, multiple sclerosis, HIV infection and chronic pain [1].
Predicting peptide properties using machine learning methods has gained interest in recent years, with a particular focus on anticancer [5], antimicrobial6 or improving permeability [7] properties, using publicly available datasets. Various methods have been proposed for prediction like support vector machines (SVM) or random forest (RF) [5]. These predictive approaches have shown interesting performance, but also limitations, especially on the type of amino acids considered, most often restricted to natural amino acids, whereas the importance of modified amino acids has been demonstrated for peptide drug design.
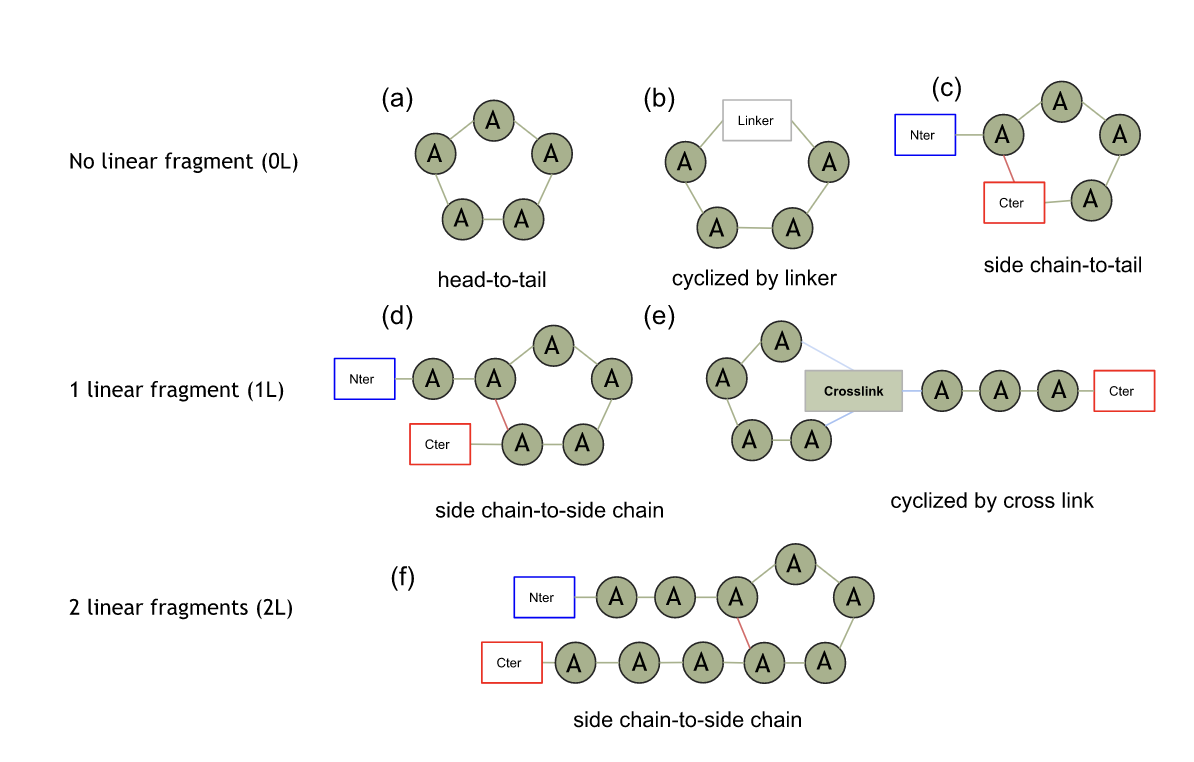
Figure 1: Simplified representation of the different types of cyclic peptides. The different graph (a) to (f) represent each a way of cyclization, while each line represent different shape of 2D graph: cycle without linear fragment (0L), cycle with 1 (1L) or with 2 linear fragments (2L). Green dots are amino acids and green links represent peptidic bonds. Red links represent side chain-to-side chain bonds and blue ones are undefined bonds. Boxes represent non-residue component (Nter or Cter extremities, linker, crosslink, etc.).
Multiple generative AI approaches for peptides have also been proposed for in silico automated peptide design. Those methods include three popular deep generative model frameworks: neural language models (NLMs), variational autoencoders (VAEs), and generative adversarial networks (GANs) [8]. However, the limitation observed in predictive modeling regarding the use of natural amino acids also applies to generative AI.
Considering this limitation, one may ask if the use of classical representation from the world of small molecules may be more relevant for peptides.
Many machine learning models for ligand-based projects on small molecules rely on vectorial representations. Two categories have been heavily used: representations based on global physico-chemical properties (LogP, TPSA, hydrogen bond donor and acceptors, molecular weight, etc.), and fingerprint representations. The latter are a widely used way to represent molecules as mathematical objects. One popular representation is the Morgan fingerprint, with great performance on virtual screening for small molecules [9].
Starting from the atomic graph of molecules (with nodes representing atoms), the Morgan algorithm takes place in two main steps: First, an identifier is assigned to each invariant (node’s characteristic) of the input graph. Then a number of iterations are performed to combine the initial node identifiers with identifiers of neighboring nodes until a specified diameter is reached. As an example, the invariants used in the Morgan algorithm to represent the nodes are the atomic number, the number of non-hydrogen neighbor atoms, the number of attached hydrogens (both implicit and explicit), the formal charge, and an additional property that indicates whether the atom is part of at least one ring.
Even if those representations have shown great performances on small molecules, they have a poor perception of the global features of molecules such as size and shape and fail to perceive structural differences which may be important in larger molecules [10].
With global molecular properties, limitations are also expected: for properties like HBD/HBA count, rotatable bond count, TPSA and logP values, predictive power may often be poor with large and flexible molecules like peptides.
Classical sequential representation used for proteins (like fasta) can help for human readability of the peptide sequence as well as for machine learning purposes. In fact, predictive and generative models have already shown some interesting performance. Yet, they were only used on natural amino acids due to the sequential representations that are limited when considering modified amino acids, or non-linear peptides.
Other peptide fingerprints for peptide prediction tasks were introduced in the literature. The Monomer Composition FingerPrints (MCFP) [11], used for the prediction of activity of non-ribosomal peptides, represents each peptide by a vector of its monomer counts. This representation can handle even cyclized peptides or peptides containing modified residues. However, it does not include any information on the sequence of residues in the peptide and on the peptide cyclizations even if this information is important for predicting peptides properties. A recent work implemented an improved version of MCFPs called Monomer Structure FingerPrints (MSFP) [12] by adding a few descriptors into the MCFP fingerprint, representing monomer clusters, 2D structures, peptide categories, and peptide diversity. MSFP performs better than MCFP, but still suffers from lack of residue sequence information.
In this work, taking into account the limitations of classical representations on peptides, we have developed new peptide fingerprints never used for predictive and generative models. Instead of using atomic graph-based representations, we developed high level graph-based representations at amino-acid and back-bone side-chain level. These representations are more intuitive and closer to the way peptides are built. They are suitable for peptides composed of natural residues, but also for complex peptides which contain modified amino-acids and cross-links.
The detection of amino acids is made using Proteax software. It parses the amino acid from a peptide and constructs a Protein Line Notation (PLN) sequence. PLN (as well as Hierarchical Editing Language for Macromolecules known as HELM [13]) are compact text representations of a peptidic chain that includes chemically significant annotations (like modified amino acids, linkers and non linearity). These languages have only been used for visual representations to our knowledge. The PLN syntax allows building an amino acid level graph, with nodes containing amino acid information, even for modified amino acid, cross linked and non-linear peptides.
This graph named Simple Peptide Graph is the basic graph representation for peptides and the most intuitive. Each node of the graph corresponds to an amino acid, and it can deal with natural and modified amino acids, cyclic peptides, crosslinks, linkers, terminal modifications, etc. An illustration is presented on Figure 2.
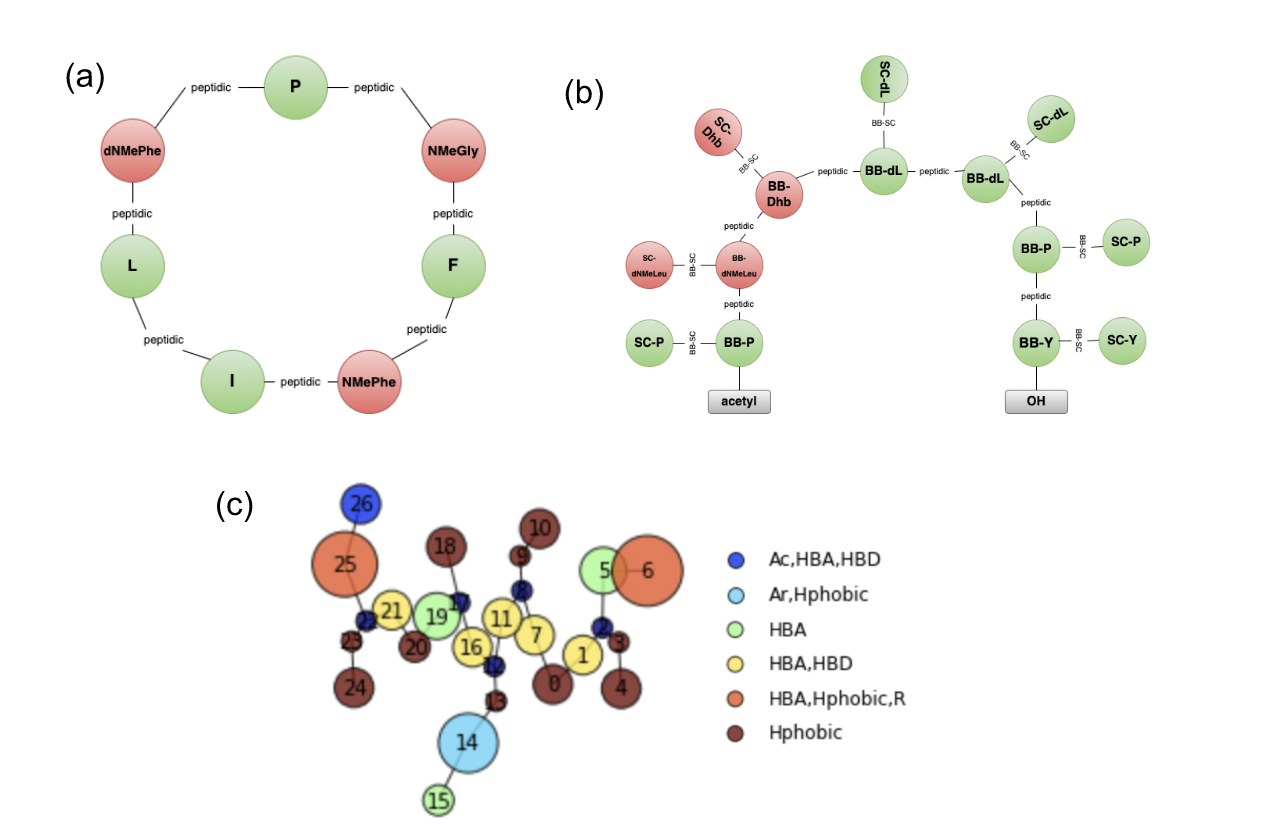
Figure 2: Peptide graph representations developed. (a) Simple peptide graph. (b) Backbone-side chain (BB-SC) peptide graph. (c) Pharmacophore representation.
We then apply a circular algorithm to transform these graphs into a vectorial representation using the Morgan algorithm. The use of circular fingerprints, encoding the presence or absence of sub-atomic structures in a vector, is therefore transposed to the amino acid graph to create a new representation.
A relevant intermediate approach is to split amino acid into backbone and side chain, so that the backbone properties are separated from the side chain, giving an interesting granularity, especially for physical chemical properties. For example, encoding N-methylation in the backbone might be more relevant than a general character for the whole amino acid.
We therefore present a second graph called BB-SC (backbone and side chain) Peptide Graph. In this graph, each amino acid node is split into a backbone node and a side-chain node as presented in Figure 2.
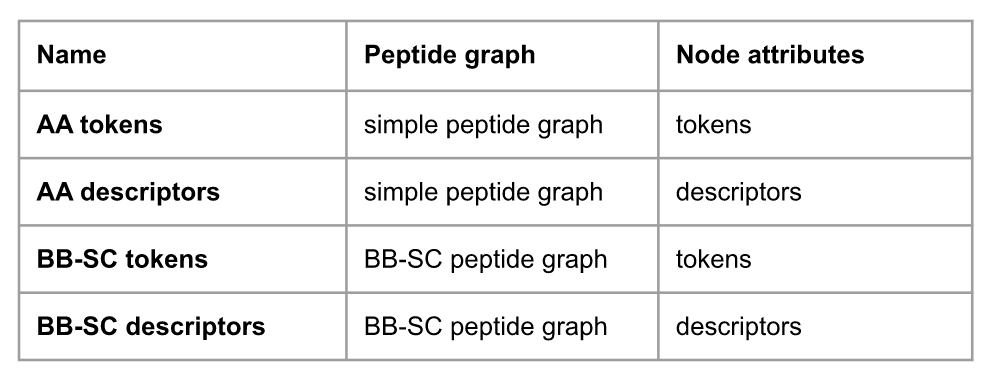
Table 1: Presentation of the different peptide fingerprints at the amino acid level with their name, the corresponding peptide graph and the chosen invariant.
For both graphs, we tested different invariants to represent the nodes: the amino acid names (tokens) or some amino acid descriptors. Given a list of descriptors with their thresholds, descriptor values are computed on each node then binned into intervals. The descriptors and number of intervals depends on the inputs.
Each type of graph combined with invariant, and using Morgan fingerprints algorithm, we could build 4 different peptide fingerprints representations. The Table 1 details the representation named corresponding to each graph and invariant.
We also implemented a representation at a fragment level, named the pharmacophore representation. The peptide is split into multiple fragment nodes following predefined functional groups like hydrophobic, HBA or HBD… The pharmacophore graph is computed from the peptide molecular graph but is at a higher level than atomic representation. This type of graph can also be used for small molecules. The pharmacophore graph is finally converted into a vectorial representation using the Morgan algorithm.
Project 1: Predictive Approach
We evaluated these new representations at the amino acid level capable of incorporating peptides with non-linearity and modified amino acids to assess whether they improve the predictive power on peptide properties compared to classical Morgan fingerprints on atomic level and global molecular descriptors for classification tasks.
We worked in collaboration with a pharmaceutical company on predicting properties of a peptide series on two targets. The objective was to be able to identify peptides achieving activity on target 1 and selectivity on target 2. The dataset was composed of 189 small linear peptides with their PIC50 measured on target 1 and target 2. Peptides included modified amino acids and other specific chemical components used to enhance peptides stability and permeability. The dataset was small, making the prediction task challenging. The distribution of peptides matching the criteria for targets 1 and 2 are presented on Figure 3, showing the dataset is balanced.
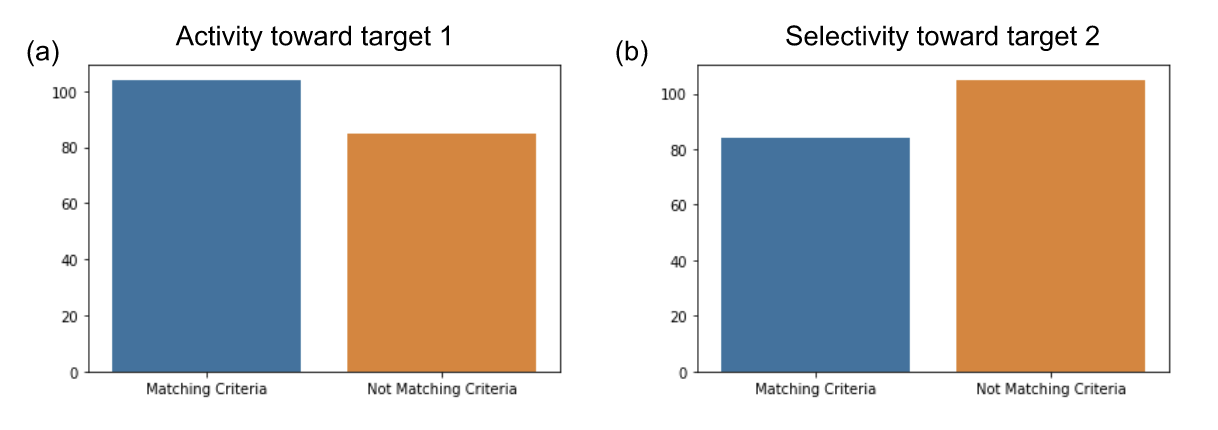
Figure 3: Peptides distribution in project 1 dataset. (a) Number of peptides matching or not the criteria for activity toward target 1. (b) Number of peptides matching or not the criteria for selectivity on target 2.
To compare the performances of each representation, multiple machine learning models, including random forest, gradient boosting and continuous logistic regression models were used. Performances given by the models were all in the same range. Therefore, only the continuous logistic regression will be present in the following. The advantage of logistic regression is that it is a simple model providing good training efficiency without a high computational cost and is also easier to implement and interpret. However, simple logistic regression also presents some disadvantages. First, the notion of order present in the initial continuous data is lost when separating active and inactive categories. There is also a threshold effect around the value of separation: considering an activity threshold with a PIC50 at 7, a peptide with a PIC50 of 6.9 will be considered inactive whereas a peptide with a PIC50 of 7.1 will be considered as active, even if they are actually quite similar.
To overcome the limitations of classification models while keeping their advantages, Iktos used a model that behaves as a classification model but takes into account the continuous information. This model behaves as a classification far from the threshold value, but as a regression locally around the threshold. Thus, the threshold effect is limited and in contrast to the simple logistic regression, there is a notion of order in the prediction allowing better correlation between target value and model prediction. However, it is still a linear model, so it may not catch complex correlations from features.
To measure the performance of our models on the dataset, we used stratified random splits as well as stratified single linkage splits. The latter are more challenging than a classical random split: they present the same distribution of active and inactive peptides in the train and test sets, and the single linkage means that we perform a clustering on peptides fingerprints. The different clusters were used for train and test sets in order to have a distribution shifted between the sets. It allows us to better evaluate the model generalization capability. We did this experiment on our 5 new representations for peptides as well as on classical Morgan fingerprints representations at the atomic level and on global descriptors at the peptide level. The performance of each model according to the split method are presented in Figure 4(a) for the activity prediction and (b) for the selectivity prediction as well as in Table 2.
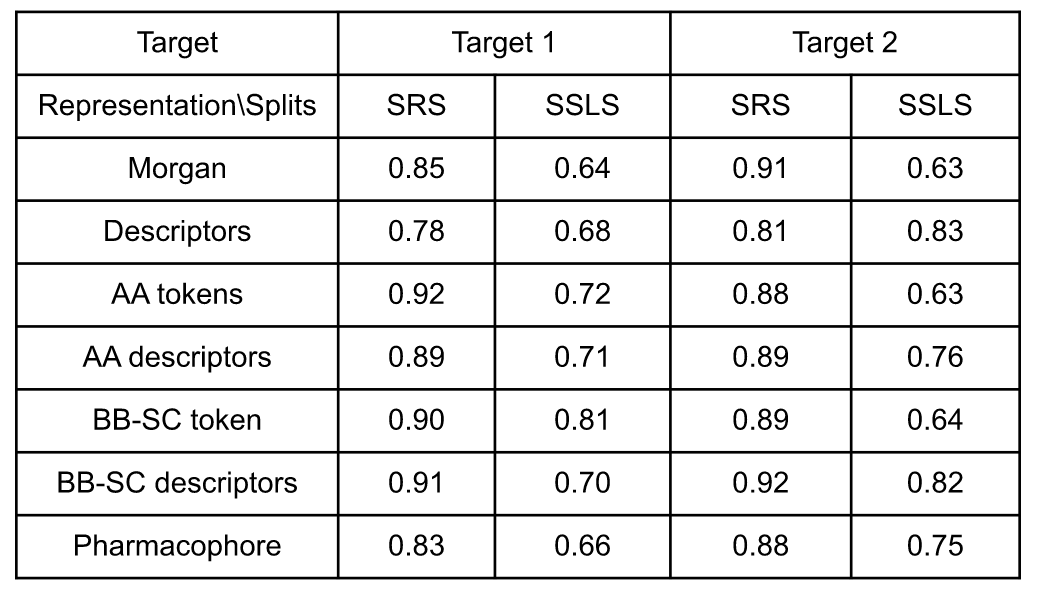
Table 2: Model performances (AUC score values) according to the representation, the split and the target. SRS stands for stratified random split and SSLS for stratified single linkage split.
For the activity prediction on target 1 with random splits, all models gave AUC scores between 0.78 and 0.92. The lowest performance was obtained by the baseline representations (0.78 for global descriptors and 0.85 for Morgan fingerprints). The amino acid level representation improved the model performance with values between 0.89 (AA descriptors) and 0.92 (AA tokens) which make a difference of 0.04 to 0.07 from the atomic Morgan fingerprints. Finally, the pharmacophore representation gives an AUC score at 0.83, lower than the amino acid level graph representation as well as atomic Morgan fingerprint.
In stratified single linkage splits, the performance of the atomic level Morgan fingerprints was lower, with an AUC score of 0.64, followed by the pharmacophore graph with an AUC score of 0.66, while the amino acid level representations have a range of values between 0.70 to 0.81. The best performance is obtained with the BB-SC token representation.
Regarding the selectivity prediction on target 2, the models perform on random splits with a range of AUC score values between 0.81 and 0.92. The global descriptors have the lowest performance while the Morgan, the pharmacophore and all amino acids level representations have similar performances ranging between 0.87 and 0.92, the best being BB-SC descriptors, followed by the Morgan (0.91).
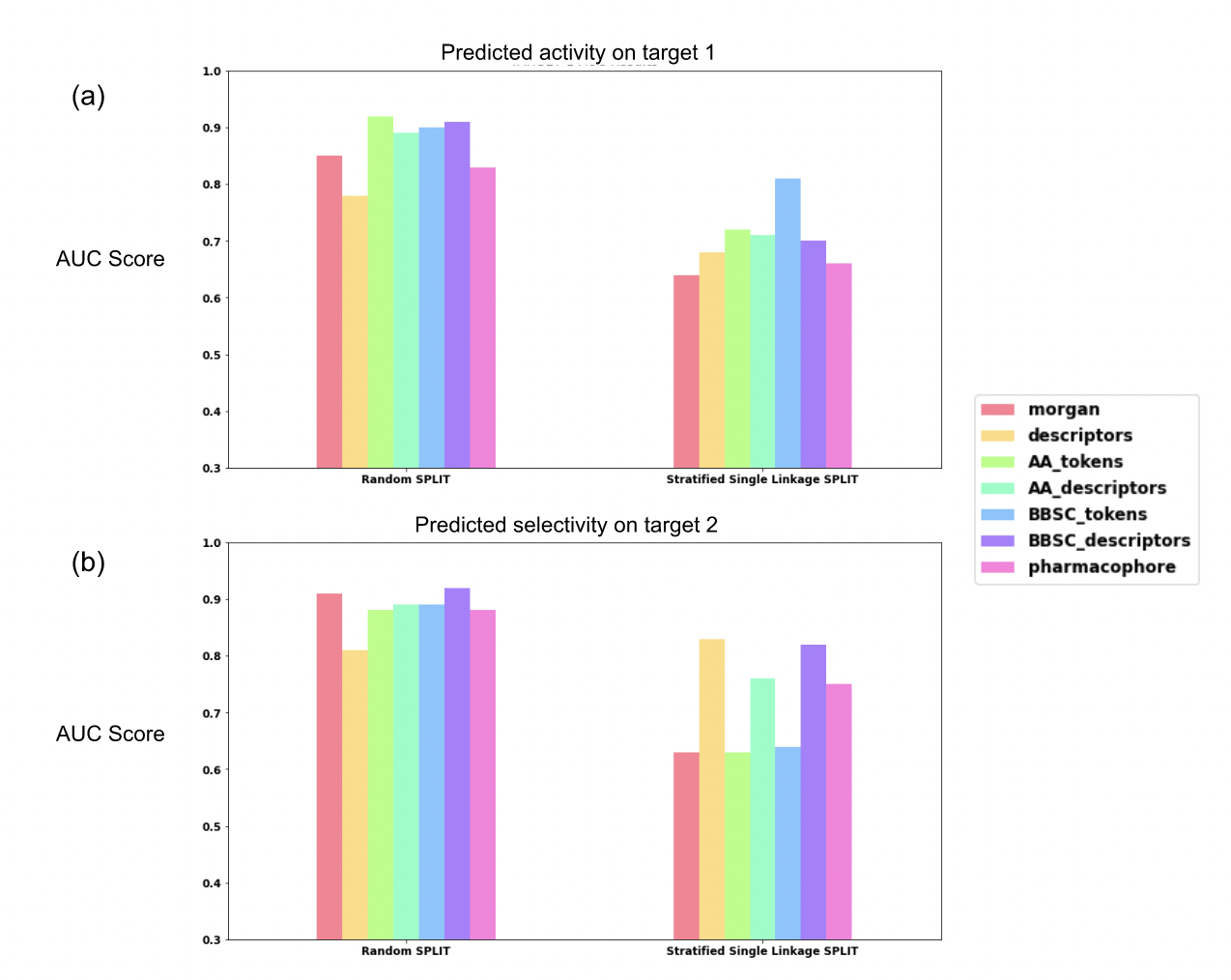
Figure 4: Prediction results (random forest). (a) AUC scores for activity prediction on target 1. (b) AUC scores for selectivity prediction on target 2.
Regarding the stratified single linkage splits, important differences in performance were observed between token-based fingerprints, descriptors and pharmacophore representations. Token-based representations, including classical Morgan, AA tokens and BB-SC tokens show low performance with AUC score values 0.63, 0.63 and 0.64 respectively. Descriptors based representations, as well as pharmacophore representation show improved performance from 0.75 to 0.83, with the highest performance shown by the global descriptors and the BB-SC descriptors representations.
Project 2: Permeability Prediction
We worked in collaboration with a second pharmaceutical company on predicting peptide permeability. They proposed a dataset composed of 5339 peptides with their measured permeability value in PIC50. The peptides from this second dataset include modified amino acids. The peptide length among the dataset is variable as presented in Figure 5.
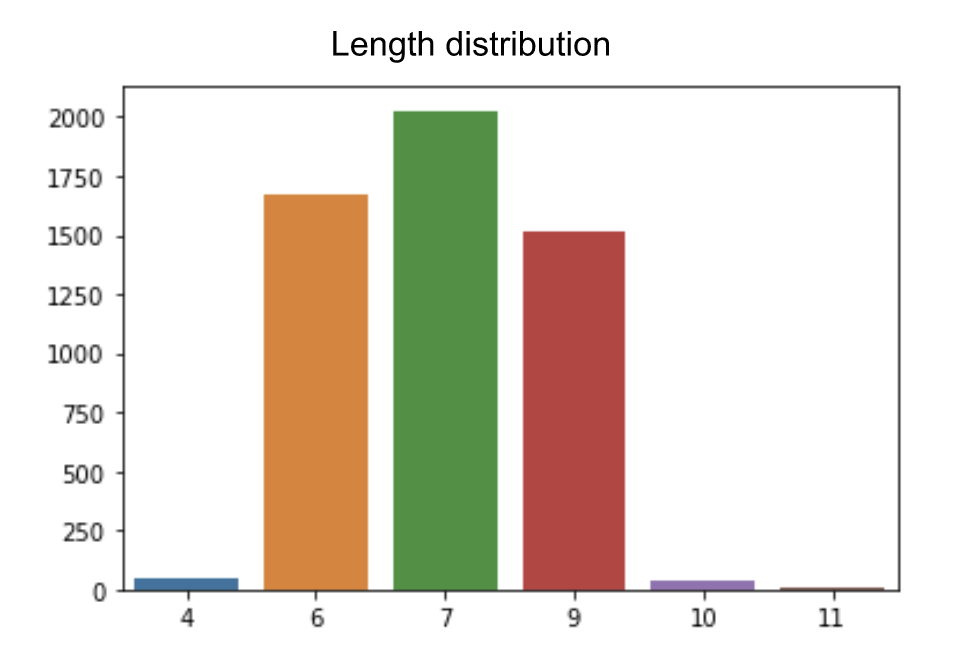
Figure 5: Peptides length distribution in the permeability dataset from project 2.
The methods used to predict permeability are the same as the ones used in the first project: we made a continuous logistic regression on the 2 baseline representations (Morgan fingerprints and global physical chemical descriptors), the 4 amino acids representations proposed in Table 1 and the pharmacophore representation. We used different splits to train the models: a stratified random split on the whole dataset and some splits based on the length or the type of the peptides. Notably, we discriminate the small peptides (≤ 6 amino acids) from the large peptides (> 6 amino acids). We also distinguished the head-to-tail cyclic peptides (0L) and the cyclic peptides with one linear fragment (1L – see Figure 1). Those different experiments were made to test the capability to generalize the prediction between two different types of peptides.
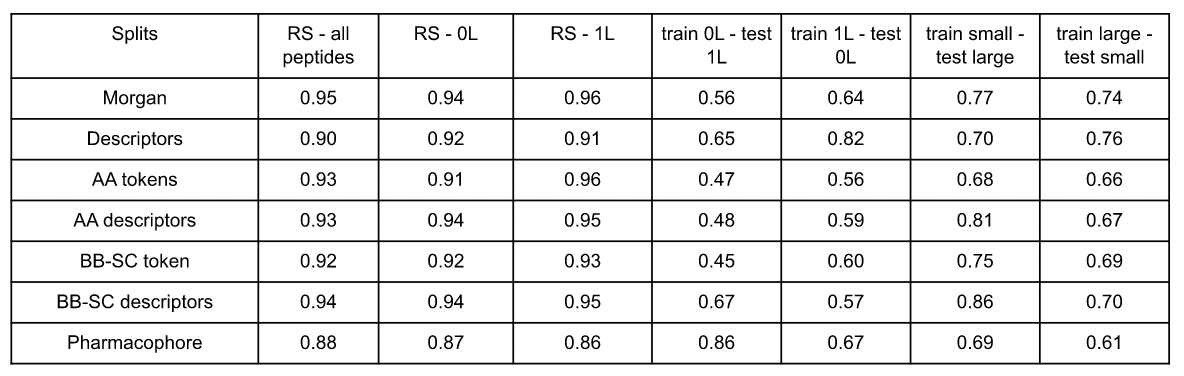
Table 3: Models performances (AUC score values) according to the representation and the split. RS stands for random split, 0L for head-to-tail cyclic peptides and 1L for cyclic peptides with 1 linear fragment.
Results of the different experiments, available in Figure 6, show that with a random split on all the peptides, all models perform with AUC value score within the same range around 0.9. The values of each model are presented in Table 3. The pharmacophore is the model presenting the lowest AUC score 0.88, while the best is the atomic Morgan fingerprints with a value of 0.95, followed closely by the AA descriptors, the BB-SC tokens and the AA tokens.
The same experiment was performed only on head-to-tail cyclic peptides and only on cyclic peptides with 1 linear fragment, giving in both cases results equivalent to the random split on all peptides.
With the models trained on head-to-tail cyclic peptides, predicting cyclic peptides with 1 linear fragment permeability gave AUC score values lower than 0.60, except for 3 of them. The BB-SC descriptors representation as well as the global descriptors have an AUC score at 0.67 and 0.65 respectively. Finally, the pharmacophore representation outperforms the other model with an AUC score at 0.86.
Training the models on cyclic peptides with 1 linear fragment to predict the head-to-tail cyclic ones gives improved performance, but all the models stay below to 0.7 of AUC score except for the global descriptors that have an AUC of 0.82. Pharmacophore representation and atomic Morgan fingerprint perform with an AUC score of 0.67 and 0.64, respectively. The amino acid level representations give lower performance between 0.56 to 0.60 of AUC score.
The prediction of large peptide properties when training models on small ones gives performances varying from 0.68 to 0.86. The BB-SC descriptor gives the best results with an AUC score of 0.86. The Morgan fingerprint performs at 0.77 while the global descriptors, the pharmacophore and the AA token representations give AUC scores at 0.70, 0.69 and 0.68 respectively.
On the other hand, the training on large peptides for small peptide properties prediction gives results with AUC score values present in the range of 0.61 to 0.76. The higher performances are given by the 2 baseline representations (atomic Morgan fingerprint and global descriptors). The lowest performance is obtained with the pharmacophore representation with an AUC score of 0.61 while all the amino acid level representations give intermediate results in the range of 0.66 to 0.70.
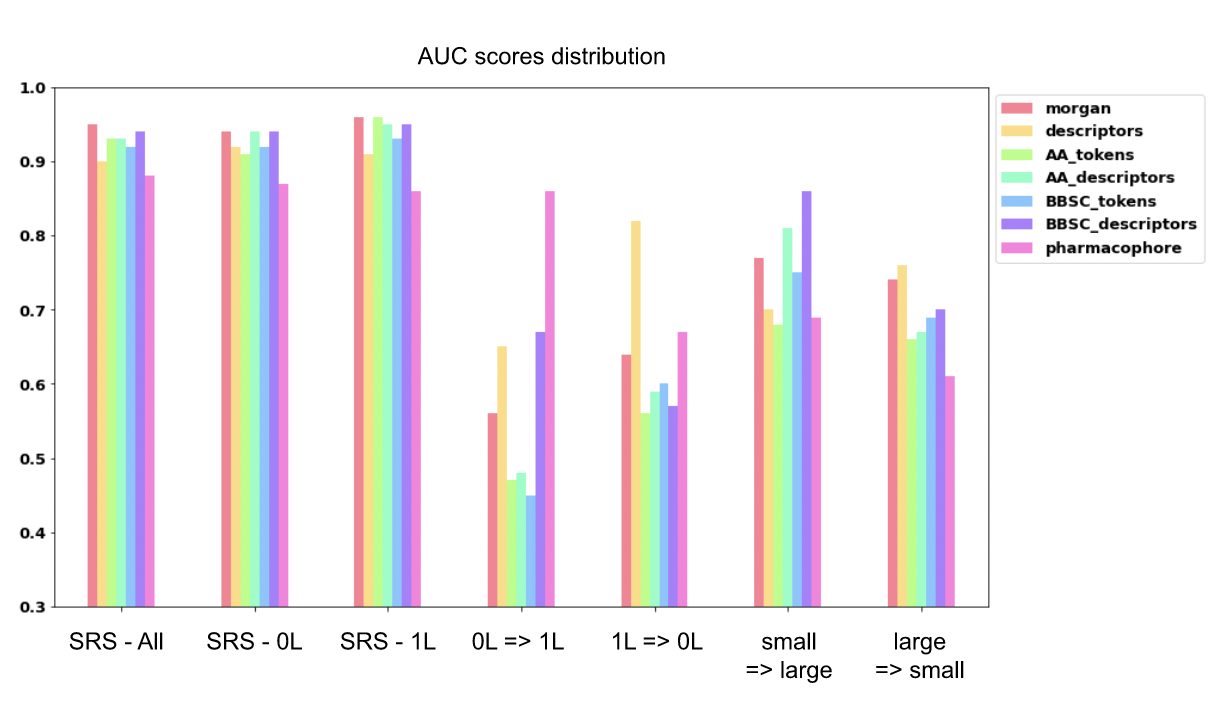
Figure 6: AUC scores for permeability prediction on different train/test splits. RS stands for random split, 0L is for cyclic peptides with no linear fragment, 1L for cyclic peptides with 1 linear fragment. The different splits are from the left to the right: SRS on all the dataset, SRS on the cyclic 0L only, SRS on the cyclic 1L only, train on the cyclic 0L and test on the cyclic 1L, train on the cyclic 1L and test on the cyclic 0L, train on small peptides and test on large peptides, and finally train on the large peptides and test on the small ones.
Project 3: Generation of Peptides
Finally, we proposed generative models based on the new amino acid level representations we developed. We were also able to design new peptides, containing modified amino acids with optimized properties.
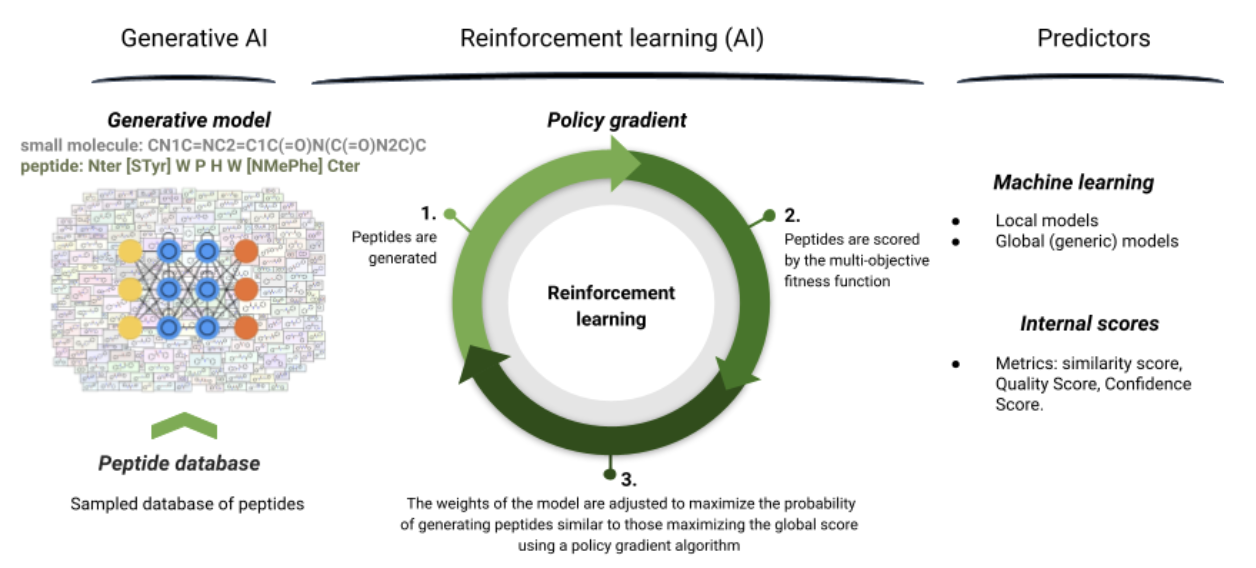
Figure 7: Pipeline for peptide LSTM generation optimized with reinforcement learning.
Iktos has developed proprietary deep learning generative modeling technology that enables the design of novel compounds meeting a predefined set of objectives in a drug discovery project. Iktos proposes AI-enabled in silico design services across the drug discovery value chain, from hit-to-lead design to late-stage lead optimization. We have successfully used our approach in various projects for small molecules, both with ligand and structure-based drug design.
In this work, we adapted our ligand-based generative technology from small molecules to peptides. The whole generative pipeline is detailed in Figure 7 and described as below: it uses a LSTM generation optimized with reinforcement learning. The pipeline is made of three main parts:
- The generative AI: in order to create a generator of peptides, we need to learn the peptide syntax on a database of peptides. To create this database, a large database of peptides is sampled randomly, relying on the initial dataset of peptide structure and residues. This database is then used to train the prior.
The prior phase allows the generator to learn the syntax and structure of desired peptides. After this step, the generator is fine-tuned on the initial dataset to be closer from the initial data distribution. It is then able to correctly sample peptides that are close to initial peptides data. - The reinforcement learning: the generated peptides are scored by a multi-objective reward function (see predictors). The weights of the model are then adjusted to maximize those scores. The aim is to increase the probability to generate new peptides close to those already generated having the highest global score. This is made using a policy gradient. This procedure is iterative and will be done for each step of the generation.
- The predictors: to guide the generation, different internal scores can be optimized depending on the project needs, like for example, the target activity predicted score (from the trained potency predictor) but also the similarity to the initial dataset and the quality of generated peptides. We also implemented an intellectual property (IP) space score defined with the ratio of amino acids that differ from the IP space of known patents. A list of checkers can also be applied to filter undesired peptides, either during the generation or after as post processing. For instance, we developed a synthesizability checker which evaluates the maximum number of violated rules for synthesizability, a solubility checker evaluating the maximum number of violated solubility rules, but also a length checker and a pattern checker. Other checkers could be applied regarding the needs of the project.
This pipeline was evaluated in collaboration with a pharmaceutical company. The aim of the project was to generate some active peptides for a desired target. They also gave us some recommendations of what a relevant peptide for the project is (regarding the synthesizability access and the solubility for example). To do so, an activity predictor was built from a dataset composed of 504 medium to large peptides, with their measured potency in PIC50. Among them, peptides include modified residues, modified terminals and crosslinks. As described in Figure 8 (a), the dataset contains mainly linear peptides but also some examples of cyclic peptides with 1 or 2 linear fragments (1L and 2L respectively – see Figure 1).
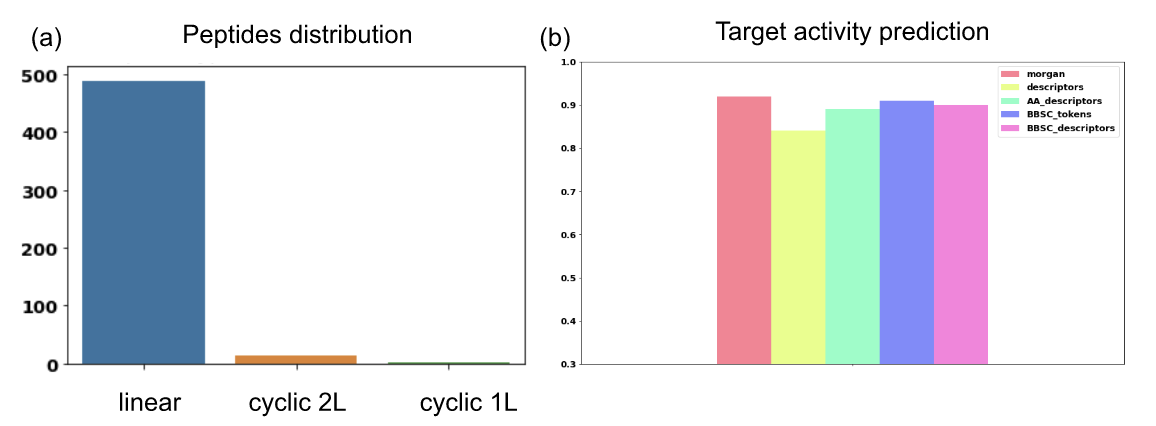
Figure 8: Properties of initial dataset used in project 3. (a) Peptide type distribution among linear, cyclic with one linear fragment (1L) and cyclic with 2 linear fragments (2L). (b) Activity prediction performances according to the representation used measure in AUC score.
The predictors on random splits gave AUC score values between 0.84 (for the global descriptors) and 0.92 (for the Morgan at the atomic level) as shown in Figure 8(b) on all models. The best model was then used during the generation to predict the potency of generated peptides.
A generation was launched using the following configuration: as scores for the multi parameter optimization, we used target activity predicted score (from the trained potency predictor), the similarity and quality scores and the IP space score previously defined. We also used a list of defined checkers to answer some wishes of the collaborator like the isoelectric point checker, the solubility checker (≤ 2 violated rules) and the synthesizability checker (≤ 3 violated rules).
Using these parameters and after 1500 steps of reinforcement learning, we generate 62,000 peptides satisfying the constraints, with a target activity prediction superior to 0.8.
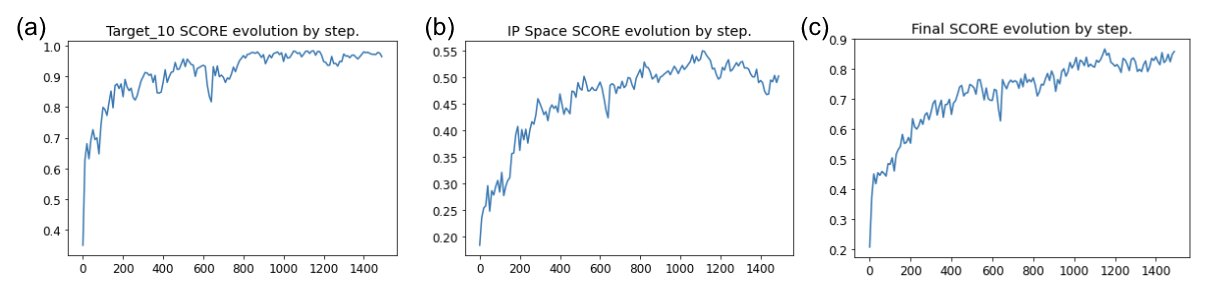
Figure 9: Evolution of different scores by steps of optimization during the peptide generation. All scores are normalized between 0 and 1 and presented as a function of the number of steps of generation. (a) predicted activity value on target. (b) IP space score value. (c) final generative score.
Figure 9 represents the evolution of the different scores that the generator tries to optimize during the iterative process of reinforcement learning. The different scores all start at 0 and increase rapidly at the beginning of the generation, meaning the generator is learning and improving its proposition. The score generally ends up reaching a plateau as, for example, the potency score stays around a score of 0.95 at the end of the generation.
Figure 10 also presents the distribution of the generated peptides according to some properties used by checkers during the generation, like the isoelectric point of the number of violated rules of synthesizability or solubility.
Figure 11 shows the distribution of the most frequent amino acid (a) in the active peptide of the initial dataset and (b) in the generated peptides.
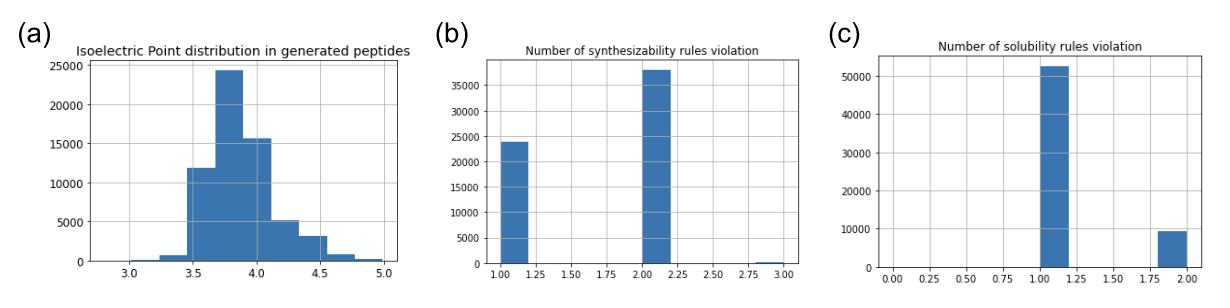
Figure 10: Checkers distribution in generated peptides. (a) isoelectric point distribution among the generated peptides. (b) number of synthesizability rules violation. (c) number of solubility rules violation.
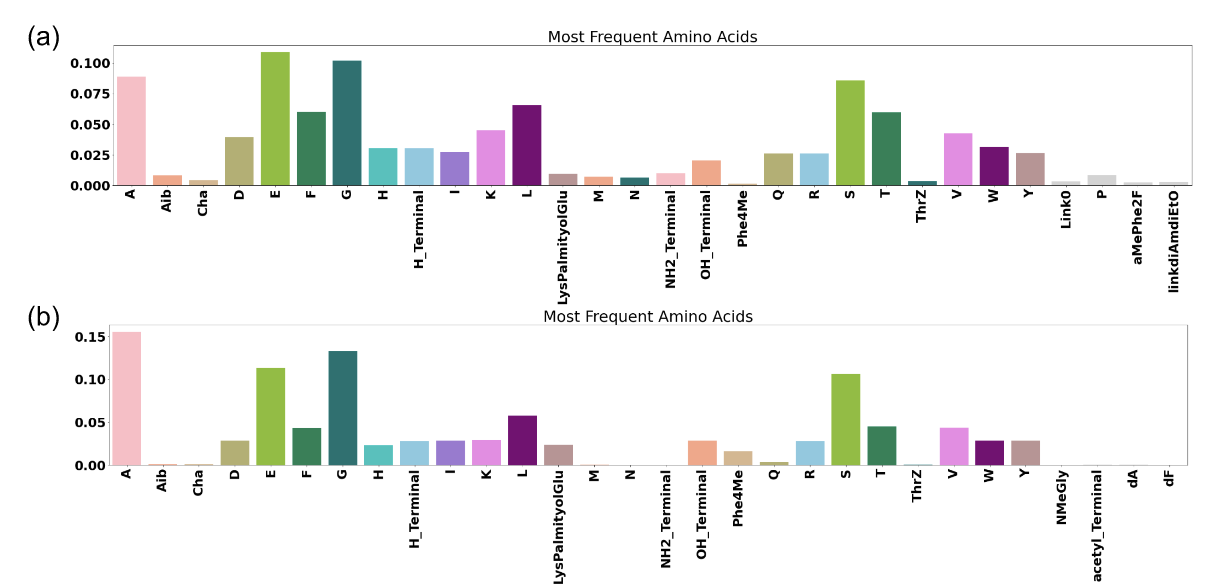
Figure 11: Distribution in amino acid in the (a) initial dataset (actives peptides only) and (b) the generated peptides. Only the most frequent amino acid are represented.
In order to visualize and evaluate the generated peptides more precisely, different visual representations have been implemented or used. Among them, we build some logos inspired from the HMM logos, well known for protein sequence comparison [14]. As biologists are familiar with this representation, it is relevant to represent an ensemble of peptides with it. The amino acid (AA) logos can describe the amino acid frequency (like presented previously in Figure 12) but incorporating the sequence position. It is particularly relevant for linear peptides of the same length. A random example of an AA logo is presented in Figure 12(a). Incorporating modified amino acids is currently feasible by two ways: using string characters other than those from the classical amino acid 1 letter code (like “Z”, “?”, “+”). It remains challenging as the number of symbols are limited and the understanding of such a code could be complex and may change from one project to another. Another way would be to cluster the non-natural amino acids, computing their distance from the natural ones and only representing the letter corresponding to the natural amino acid of the clusters in the logo. Finally, we developed another interesting way to represent the peptide properties with a logo, even with modified amino acids, but still being easily understandable, as illustrated in Figure 12(b). This pharmacophoric (PH) logo can illustrate a property of the amino acids (like the hydrophobicity, the positive charge or the polarity) at a given position of the peptide sequence.
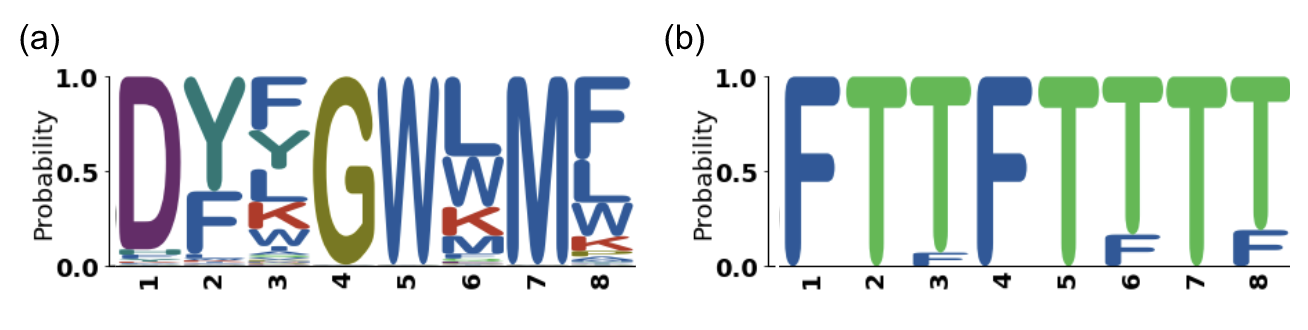
Figure 12: Logos inspired from HMM logos. (a) AA logo for amino acids sequence. Letters represent the one letter code of amino acids while color represents the amino acid propertie (red is positively charged amino acid for example). (b) PH logo for an amino acid property. Here it represent the amino acid polarity. Letters are F: false and T: true meaning nonpolar and polar respectively.
An important property to evaluate the generated peptides is the diversity. To understand how similar or not the generated peptides are from each other and help the selection of the most promising compounds, a clustering can be made. An example of clustering visualization on generated peptides is presented in Figure 13 where a K-means clustering was made using peptide fingerprints (AA tokens) and 100 clusters.
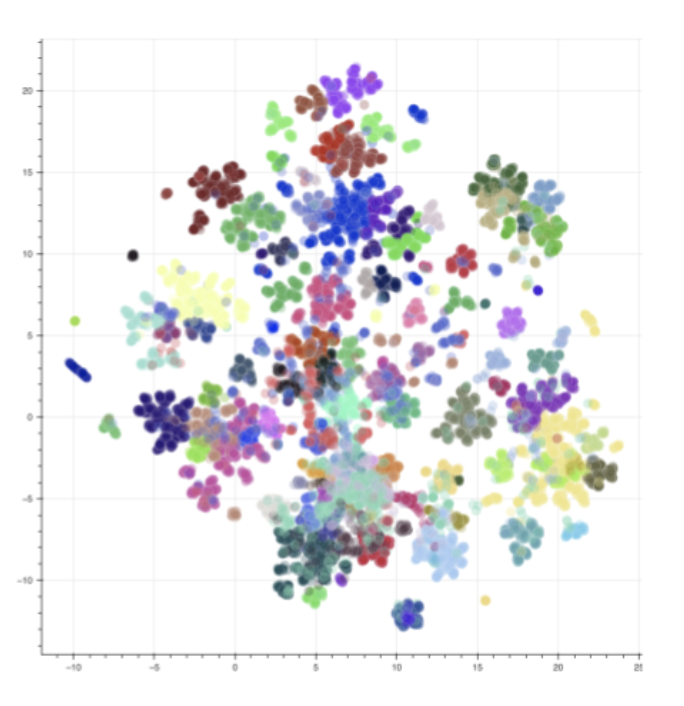
Figure 13: Visualisation of peptides clustering with UMAP. Clusters were made with K-means and each color corresponds to a cluster.
The goal of the clustering is to recover n=100 diverse peptides from generation and select the peptide maximizing the scores on each cluster.
The clusters were then visualized based on a uniform manifold approximation and projection (UMAP) [15]. UMAP is a dimension reduction technique that can be used to visualize the chemical space. In the figure, each color represents a cluster. Another relevant representation to easily understand the diversity of peptides and what modifications have been proposed by the generator is the use of a sequence alignment. Merck has recently developed a free and open-source package named PepSeA [16] for visualization and alignment of peptide sequences. This tool is capable of managing the non-standard amino acid residues and non-linear peptides thanks to the use of HELM sequence. We incorporate PepSeA into our pipeline for peptide evaluation. An example of peptides alignment with PepSeA is presented in Figure 14. It notably allows to understand how homogeneous or diverse a dataset is if some parts of the sequence are conserved or not.

Figure 14: Cluster visualisation with PepSeA after alignment of the generated sequence. Each node is an amino acid. Greens are conserved amino acids, reds are changes from a peptide sequence of reference.
Discussion
In this work, we introduced new graph representations of peptides at different levels. Those graphs take into account non-natural amino acids and non-linear peptides allowing to work on real case projects for peptide-based drug design. Converted into a vectorial representation using the Morgan algorithm, we compared those new representations with the classical Morgan fingerprint on atomic graph and with global molecular descriptors on classification tasks.
Based on the 3 projects we exposed in this paper; we show promising results obtained with peptide graph representations.
For the first project, the peptide fingerprints outperform Morgan fingerprints in random split and generalizes better for target 1 as illustrated by the performance in stratified single linkage. Indeed, the Morgan representation suffered the biggest drop in AUC, consistent with its ability to perform well on homogeneous distribution, but a difficulty to generalize. The BB-SC token representation on the contrary underwent a lower decline which could be an indication of its ability to better generalize. For target 2, the peptide fingerprints are equivalent to the Morgan fingerprint in a random split and the peptide fingerprints relying on descriptor invariants have better performance in single linkage split.
For the second project, performance on random split gives the peptide fingerprints equivalent to the Morgan fingerprint. However, performance on the different splits according to the peptide size or type gave heterogeneous performance.
From a more general point of view, the performance of the models really depends on the dataset and the split used, showing how challenging the prediction on peptides is. Using random split, peptide graph representations gave performances nearly equivalent to the baseline atomic Morgan fingerprint and improved compared to the use of global descriptors. On the contrary, using stratified single linkage split, the atomic Morgan fingerprint shows the lowest performance, while peptide graph representations and global descriptors give improved performances. This shows how some representations like the atomic Morgan fingerprint have difficulties to generalize. Again, the best models depend on the dataset as we saw comparing the performance on target 1 and target 2 in project 1 or the results for project 2.
A good example would be the pharmacophore representation which often gives among the lowest performance except in project 2 for the prediction of cycloCL peptides permeability while training on cyclic peptides with a performance that outperforms the baseline and the amino acid level representations by far.
Another interesting result is the fact that the different descriptor-based representations generalize better on target 2 of project 1 than the token representations. Even if the interpretation of such results is challenging, it would be interesting to understand why the descriptors generalize better than token representations on this target. In order to help understand the impact on that, interpretability of ML models could be interesting. Yet, atomic Morgan fingerprint interpretability on peptides is limited by the too many invariants and the fact that they do not encode the amino acid sequence. Figure 15 shows an example of Morgan interpretability results using SHAP values analysis. SHAP values (SHapley Additive exPlanations) [17] is a method based on cooperative game theory, used to increase transparency and interpretability of machine learning models. In Figure 15, more than 20.000 features representing small fragments were recovered from the peptides. Interpreting the results may be challenging.
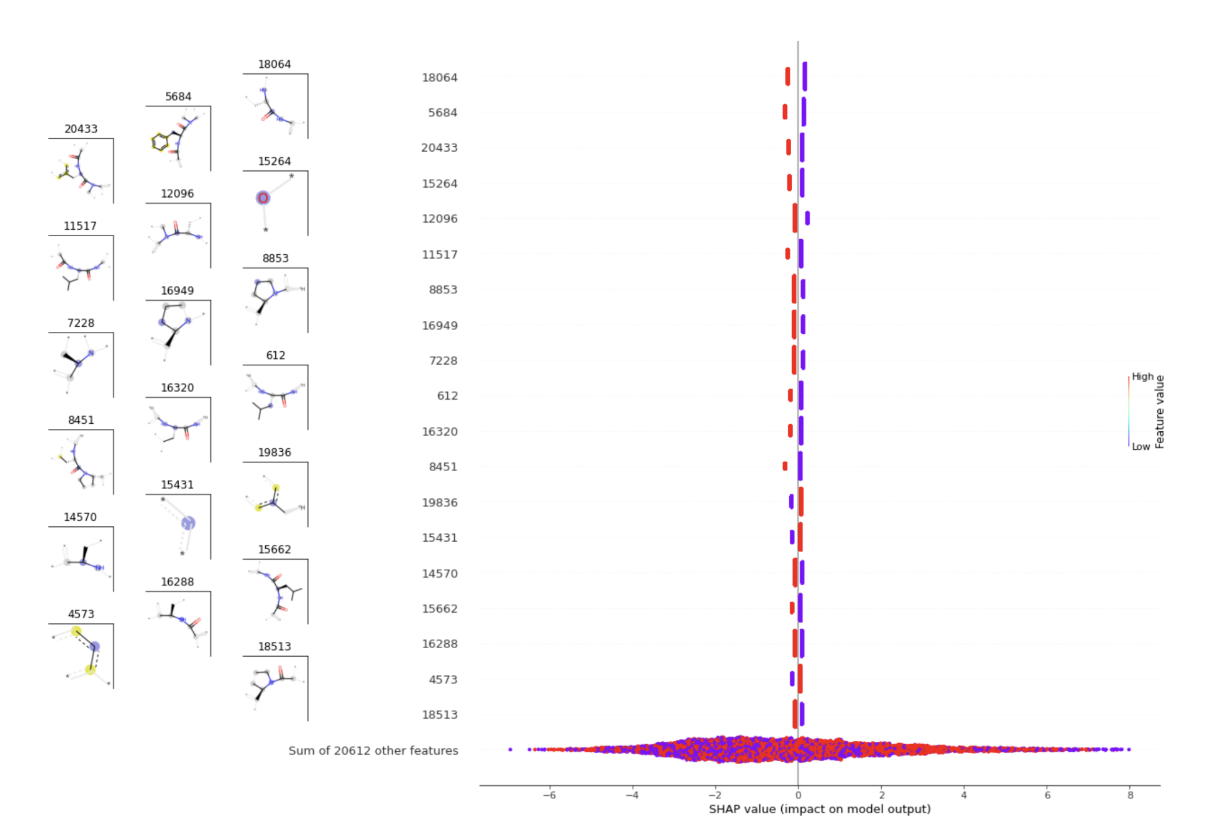
Figure 15 : Interpretability on Morgan fingerprints using SHAP values.
On the other hand, even if descriptor representation is easier to interpret, it is still limited for peptides as it considers its global properties only as in Figure 16.
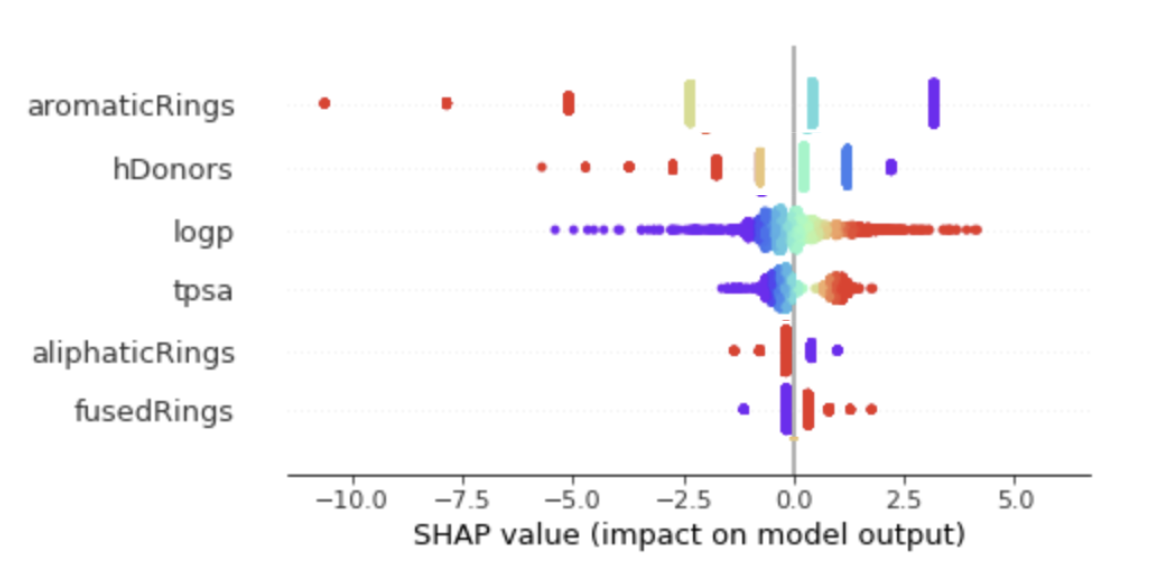
Figure 16 : Interpretability results on descriptors representation using SHAP values.
In fact, peptide amino acids-sequence patterns are the most important features to consider when analyzing peptides. Peptide fingerprints at the amino acid level have an interpretability more representative to understand important amino acid sequence patterns, giving another interesting argument in favor of peptide fingerprint representations. For example, with peptide fingerprint representation, interpretability will rely on features representing amino acid sequences as in Figure 17, that are more representative for a chemist on peptides.
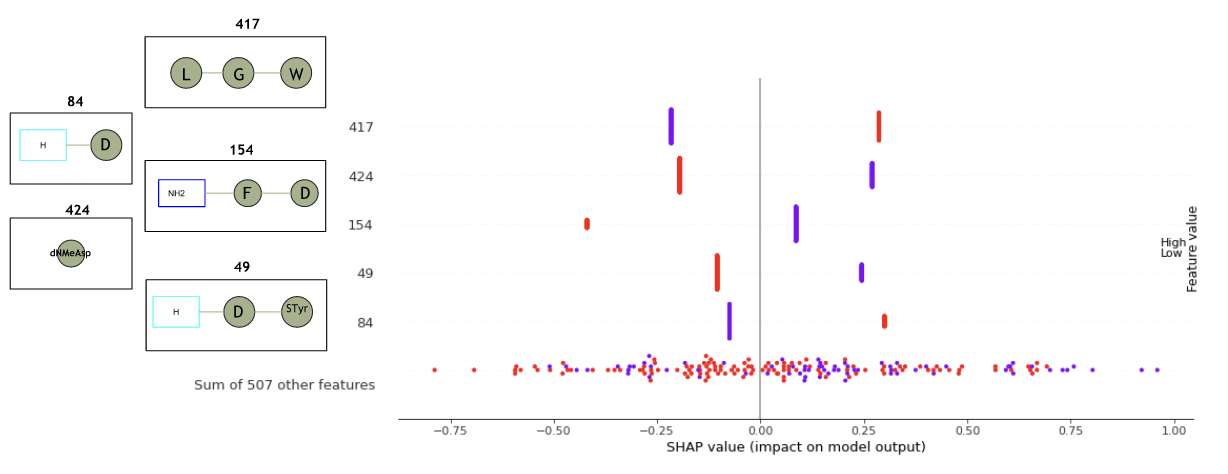
Figure 17 : Features interpretability of peptide fingerprints.
In this work, we focused on the construction of 2D descriptors and representation for peptides. Although the effort to build 3D descriptors can be significant, they can be a game-changer for the prediction of key properties of peptides. They can in particular make it possible to gain relevant information regarding flexibility, a critical aspect of permeability [18]. For this purpose, we also worked on a pipeline to sample peptides conformations to construct some 3D descriptors. Currently, we haven’t succeeded in improving our models incorporating 3D information. However, this is an active field of research for us, with some work in progress and numerous areas for improvement.
Regarding project 3, we used the amino acid level predictors to generate new peptides with optimized predicted properties. The distribution of the peptide properties used by checkers during the generation put in light that the generator respects the constraint imposed like for the maximal number of violated rules for synthesizability. Also, the distribution of amino acid between the initial dataset and the generated peptides is very similar, meaning the generator was able to learn the syntax for potent peptides.
Feedback on the generated peptides from our collaborator were positive: we succeeded in obtaining new peptides matching the optimization criteria, keeping the crucial part of the amino acid sequence for the activity (without any knowledge of this motif) and still getting some diversity. These results comfort us to continue improving our promising technology for peptide design.
Bibliography:
[1] Muttenthaler, Markus, et al. “Trends in peptide drug discovery.” Nature Reviews Drug Discovery 20.4 (2021): 309-325.
[2] Wang, Lei, et al. “Therapeutic peptides: current applications and future directions.” Signal Transduction and Targeted Therapy 7.1 (2022): 1-27
[3] Santos, Gabriela B., A. Ganesan, and Flavio S. Emery. “Oral administration of peptide‐based drugs: beyond Lipinski’s Rule.” ChemMedChem 11.20 (2016): 2245-2251.
[4] Philippe, Grégoire JB, David J. Craik, and Sónia T. Henriques. “Converting peptides into drugs targeting intracellular protein–protein interactions.” Drug Discovery Today 26.6 (2021): 1521-1531.
[5] Basith, Shaherin, et al. “Evolution of machine learning algorithms in the prediction and design of anticancer peptides.” Current Protein and Peptide Science 21.12 (2020): 1242-1250.
[6] Rondon-Villarreal, Paola, Daniel A Sierra, and Rodrigo Torres. “Machine learning in the rational design of antimicrobial peptides.” Current Computer-Aided Drug Design 10.3 (2014): 183-190.
[7] Wei, Huan-Huan, et al. “The development of machine learning methods in cell-penetrating peptides identification: a brief review.” Current Drug Metabolism 20.3 (2019): 217-223.
[8] Wan, Fangping, Daphne Kontogiorgos-Heintz, and Cesar de la Fuente-Nunez. “Deep generative models for peptide design.” Digital Discovery (2022).
[9] Capecchi, Alice, Daniel Probst, and Jean-Louis Reymond. “One molecular fingerprint to rule them all: drugs, biomolecules, and the metabolome.” Journal of cheminformatics 12.1 (2020): 1-15.
[10] Capecchi, Alice, Daniel Probst, and Jean-Louis Reymond. “One molecular fingerprint to rule them all: drugs, biomolecules, and the metabolome.” Journal of cheminformatics 12.1 (2020): 1-15.
[11] Abdo, Ammar, et al. “A new fingerprint to predict nonribosomal peptides activity.” Journal of computer-aided molecular design 26.10 (2012): 1187-1194.
[12] Abdo, Ammar, et al. “Monomer structure fingerprints: an extension of the monomer composition version for peptide databases.” Journal of Computer-Aided Molecular Design 34.11 (2020): 1147-1156.
[13] Zhang, Tianhong, et al. “HELM: a hierarchical notation language for complex biomolecule structure representation.” (2012): 2796-2806.
[14] Schuster-Böckler, Benjamin, Jörg Schultz, and Sven Rahmann. “HMM Logos for visualization of protein families.” BMC bioinformatics 5.1 (2004): 1-8.
[15] McInnes, Leland, John Healy, and James Melville. “Umap: Uniform manifold approximation and projection for dimension reduction.” arXiv preprint arXiv:1802.03426 (2018).
[16] Baylon, Javier L., et al. “PepSeA: Peptide Sequence Alignment and Visualization Tools to Enable Lead Optimization.” Journal of Chemical Information and Modeling 62.5 (2022): 1259-1267.
[17] Štrumbelj, Erik, and Igor Kononenko. “Explaining prediction models and individual predictions with feature contributions.” Knowledge and information systems 41.3 (2014): 647-665.
[18] Rezai, Taha, et al. “Conformational flexibility, internal hydrogen bonding, and passive membrane permeability: successful in silico prediction of the relative permeabilities of cyclic peptides.” Journal of the American Chemical Society 128.43 (2006): 14073-14080.
